Data collected from the affiliated PRISM sites will be aggregated at I2C where strict Q/C and secure data transfer protocols will be enacted and enforced. I2C will facilitate the pre-processing and post processing of the data and then provide it to the NIH-led NDAR database for the widest possible dissemination.
We have five specific aims and have structured our imaging and informatics workgroups around them.-
Specific Aim 1
Oversee the collection, compatibility, aggregation, and quality control of neuroimaging data obtained from the multi-site PRISM consortium.
-
Specific Aim 2
Ensure the harmonization of neuroimaging, neuropsychological, and clinical metadata using a robust database architecture.
-
Specific Aim 3
Develop and optimize standardized data processing algorithms for within and between subject longitudinal modeling.
-
Specific Aim 4
Operationalize end-to-end data analyses by linking of multi-modal PRISM data processing tools through cutting-edge scientific workflow systems and large-scale computation.
-
Specific Aim 5
Develop a dedicated project website to promote, outreach, communication, policy compliance, and system usage logging.
Specific Aim 1
Standard Neuroimaging Protocol-Imaging Workgroup (Acquisition)
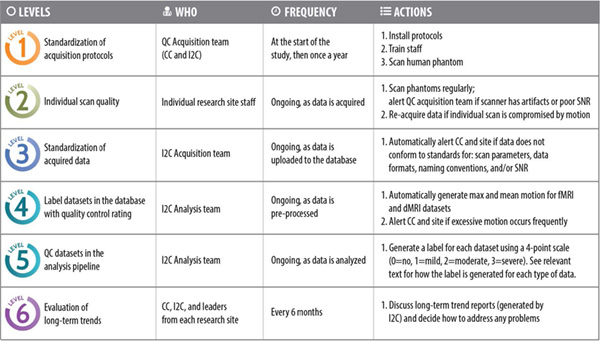
We created an optimized, cutting-edge protocol that minimizes temporal and inter-site variance in addition to motion-artifacts at the source. The required Core Protocol was modeled after the Human Connectome Lifespan Project (HCLP) and the Pediatric Imaging Neurocognitive Genetic (PING) study.
In multi-site, longitudinal studies, consistency in data collection practices is key. Short-term and long-term trends in data quality will be assessed within and across the collection sites and each site will be responsible for monitoring data quality and alerting the MR team as problems arise.
To ensure the highest level of quality control, a six level program has been designed.
Specific Aim 2
Databasing-Imaging & Informatics Workgroups
An ideal database doesn't just store information-it facilitates the extraction of the maximum amount of knowledge from the acquired data. To ensure the effectiveness of I2C's databases, three main ideals are adhered to:
- Quality of data uploaded to form the database.
- Organization of data within the database.
- Ease of extracting data.
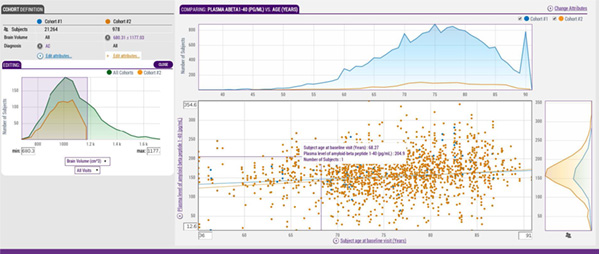
This ensures that the data is organized in a consistent way and doesn't require that all who are seeking the information be a "power user" of the database environment. At LONI, we developed LONI QC, an automated interface for quality control that allows users to set criteria for image quality during the download process.
LONI also has the Image and Data Archive (IDA), an easy-to-use platform-independent system designed to handle large-scale archiving of multi-site neuroimaging data.
We have many years of experience mapping data from consortia-specific schemas into a common model and are developing powerful semi-automated schema mapping tools that will manage the effort of mapping from the ABCD repository into National Database for Autism Research (NDAR). Data stored in the LONI IDA is exported, prepared, and submitted to NDAR and we have developed direct data processing interoperability between the LONI Pipeline and NDAR, which is particularly relevant to our data analytic activities under this multi-site project.
Specific Aim 3
Standardized Processing-Imaging Workgroup (Analysis)
Procedures for processing each type of imaging data will be developed within the imaging workgroup. The informatics workgroup will oversee the transfer of the data in and out of the database for processing to ensure that no errors are introduced. The database will release processed data and pipelines used to generate the processed data.Each of the following will have their own dedicated protocol:
- Standardized Minimal Preprocessing
- Anatomical Processing and Modeling
- Structural MRI (sMRI)
- Automated Cerebellar Partitioning
- Diffusion MRI (dMRI)
- fMRI Analyses
- fLongitudinal fMRI Analysis
Specific Aim 4
Data Analyses with Workflow Systems and Large-Scale Computation-Imaging Analysis and Informatics Workgroups
I2C will address the aims set by the PRISM project by deploying specific data processing streams, statistical comparisons and analyses, and methods for visualization of results. The LONI Interrogator will be used to explore basic relationships among the collected datatypes and leverage data mining tools to generate additional hypotheses.
Streamlined scientific workflows will be created using the LONI Pipeline infrastructure to create and provide a natural data provenance. The LONI Pipeline is an effective tool that allows for the integration of disparate resources and effortlessly disseminates resource metadata descriptions using web-based services.
Modules, workflows and data provenance in this project will streamline data access, use, and management, which will aid the user-focused concepts and core design of this project.
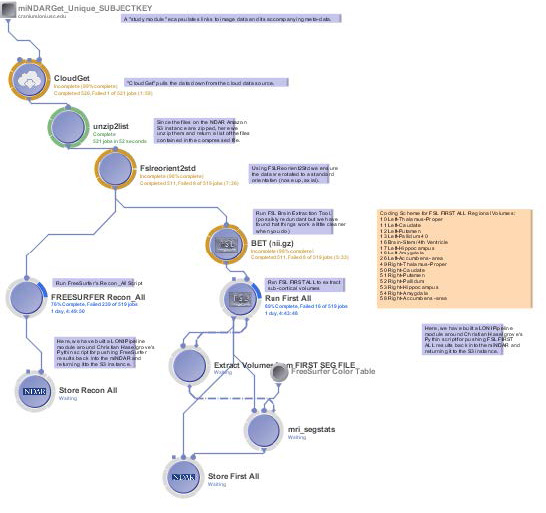
In addition, a stringent data processing algorithm and workflow validation plan will be used to ensure the proper validation of data. Automated segmentation will use a test atlas set and the validity of the results will be evaluated via automated and manual comparisons.
Specific Aim 5
Communication and Monitoring-Informatics Workgroup
I2C will oversee the curation and moderation of meaningful and instructional web content to create a communication hub for potential subjects, participants, the PRISM consortium, the NIH, and the scientific community at large.This site will provide valuable resources in the forms of data, tools, publications, tutorials and general information. It will also be the centralized location for all questions regarding data acquisition and processing for the PRISM project.